top of page

How do transcription factor hubs facilitate target search and occupancy?
Project Leads: Net Ratchasanmuang, Dr. Apratim Mukherjee, S Fallacaro
How transcription factors efficiently find and bind their targets in the crowded nuclear environment is poorly understood. We have previously shown (Genes and Dev, 2017 and eLife, 2018) that transient high-local concentration "hubs" formed by the pioneer factor Zelda and it's co-activators enable robust target gene occupancy and activation. We are now using mutant forms of Zelda along with live-imaging, single-molecule tracking, and genomics to understand the role of DNA binding vs. Protein-protein interactions in hub formation and to elucidate how hubs facilitate the transcription factor search process.
Video: Zelda (green) and Chromatin (white) during early nuclear divisions in a Drosophila embryo
Nuclear Compartmentalization and Dynamics During
Zygotic Genome Activation
Project Lead: Dr. Apratim Mukherjee
Nuclear compartmentalization and chromatin organization are established as an organism undergoes the maternal to zygotic transition (MZT). How these emerging nuclear patterns modulate the kinetics of transcription factors and key transcriptional machinery proteins such as RNA Polymerase II remains poorly understood. I will explore this question using a combination of single molecule tracking and rapid, volumetric imaging in the context of a live, developing Drosophila embryo.
Video: Heterochromatin formation during Zygotic Genome Activation in Drosophila visualized by imaging HP1 GFP
Video: Single molecule tracking of RNA Polymerase II at 100 frames/ second in a Drosophila embryo
Transcription Factor Hub Formation and Function
Project Lead: S Fallacaro
Many transcription factors organize into highly dynamic hubs in eukaryotic nuclei. Hubs are thought to impact transcription by modulating transcription factor interactions with chromatin and selectively partitioning cofactors. My goal is to study how hubs form at individual gene loci and assess how the variety in hub formation leads to differential transcriptional outcomes.
Video: Dorsal (green) hubs in nuclei of early Drosophila embryos with transcription at a Dorsal target gene (Sna, magenta).
Transcriptional repression and chromatin dynamics
Project Lead: Ella Hayward-Lara
Transcriptional repressors bind to chromatin on very short timescales but must mediate extraordinarily stable repressive outcomes to avoid pathogenic mis-activation of genes. For example, Polycomb repression establishes tissue identity in the embryo by repressing key developmental genes, and actively maintains this repression for the lifetime of an organism. Polycomb proteins are enriched in Polycomb bodies, which are relatively stable hub-like structures that are essential for repression and are formed as repression is established in the embryo. My project aims to uncover how the biophysical properties of Polycomb bodies could mediate changes to repressed chromatin and lead to stable repressive outcomes.
Video: Tracking the dynamics of two active gene loci in one nucleus inside an early Drosophila embryo
Molecular kinetics of transcriptional regulators and machinery
Project Lead: Dr. Santosh Adhikari
Fundamental biological processes such as transcription is regulated by highly dynamic molecular scale events involving multiple molecules that are organized within nuclear microenvironments. Live single-molecule imaging can map the spatial organization as well as the kinetics of regulatory molecules within these microenvironments with high specificity and single molecule sensitivity with ~30 nm resolution. My project involves developing novel single molecule imaging and analysis approaches to understand the biophysical principle governing the dynamics of these microenvironments and how they modulate the kinetics of individual molecules to regulate the transcription.
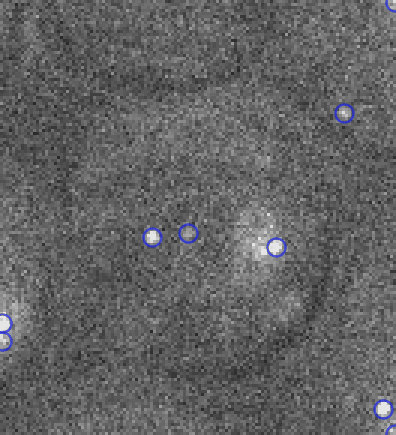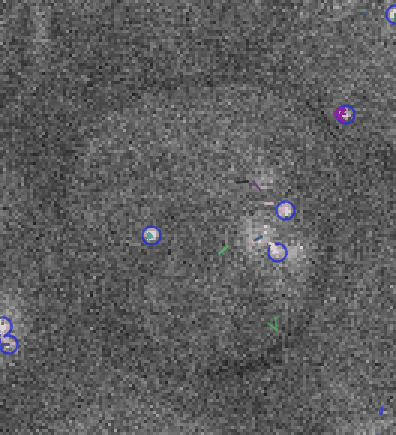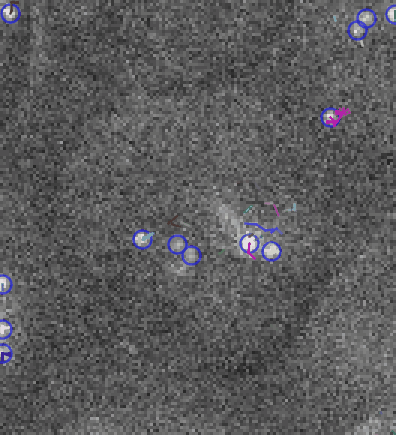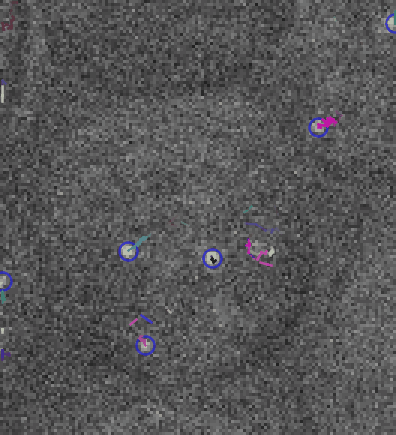
Video: Single molecule tracking of RNA Polymerase I in nucleoli
Oncogenic hubs formed by ENL
Project Lead: Kaeli Mathias (co-mentored by Prof. Liling Wan)
Aberrant transcriptional hub formation, defined by high local concentrations of proteins, has been associated with disease, including cancers and human expansion repeat disease, yet the exact functional consequence of hub formation remains unknown. Oncogenic mutations in the chromatin reader protein, ENL, are linked to transcriptional hub formation and are required for hyper-transcriptional activation of target genes. The goal of my project is to use oncogenic mutations found in the chromatin reader protein, ENL and a combination of advanced imaging techniques, including single molecule tracking and live imaging of transcription, to determine the effect of hub formation on the molecular kinetics of incorporated proteins and transcription dynamics.
Video: Aberrant hubs formed by ENL mutants in HEK 293 Cells
bottom of page
